Molecular Plasmonics: Nanoparticle-based applications for life science and catalysis
Ekaterina Podlesnaia, Stephan Kastner, Lisa Stolle, Nicole Slesiona, Andre Heewig, Cornelia Reuter, Anne-Kathrin Dietel, Andrea Csáki and Wolfgang Fritzsche
Department of Nano Biophotonics, Leibniz Institute of Photonic Technologies, Jena, Germany
The research work focuses on the potential of optical effects on plasmonic nanostructures. The design and synthesis of metal nanoparticles and nanostructures with desired defined optical properties (localized surface plasmon resonance, LSPR) in combination with (bio)molecular components (e.g. DNA) forms the technological basis. In particular, shape anisotropic plasmonic nanoparticles are precisely produced with controlled multistep and/or microfluidic synthesis [1]. After bioconjugation, the generated functional nanostructures allow applications as optical markers and sensors [2]. In sensor technology or sensory bioassays (colorimetry), plasmonic nanostructures act as optical signal converters, also in a multiplexed (microarray) format, with customized imaging spectrometric readout approaches [3, 4, 5]. Such passive plasmonic nanostructures allow applications in medical diagnostics, food and water analysis, and environmental technology [3]. On the other hand, active nanostructures convert energy acting as optical antennas, which can be used for applications in catalysis and materials processing [6].
a) Microfluidically synthetized plasmonic nanoparticles. b) Gold nanocube arranged on DNA-origami. c) colorimetric DNA-assay and miniaturized DNA amplification device
References:
[1] Podlesnaia, E.; Csáki, A.; Fritzsche, W., Time Optimization of Seed-Mediated Gold Nanotriangle Synthesis Based on Kinetic Studies. Nanomaterials 2021, 11 (4), 1049.
[2] Slesiona, N.; Thamm, S.; Stolle, H. L. K. S., et al., DNA-Biofunctionalization of CTAC-Capped Gold Nanocubes. Nanomaterials 2020, 10 (6), 1119.
[3] Reuter, C.; Urban, M.; Arnold, M., et al., 2-LED-μSpectrophotometer for Rapid On-Site Detection of Pathogens Using Noble-Metal Nanoparticle-Based Colorimetric Assays. Applied Sciences 2020, 10 (8), 2658.
[4] Barbir, R.; Pem, B.; Kalčec, N., et al., Application of Localized Surface Plasmon Resonance Spectroscopy to Investigate a Nano–Bio Interface. Langmuir 2021.
[5] Zopf, D.; Pittner, A.; Dathe, A., et al., Plasmonic Nanosensor Array for Multiplexed DNA-based Pathogen Detection. ACS Sensors 2019, 4 (2), 335-343.
[6] Stolle, H.; Csáki, A.; Dellith, J., et al., Modification of Surface Bond Au Nanospheres by Chemically and Plasmonically Induced Pd Deposition. Nanomaterials (Basel) 2021, 11 (1).
Acknowledgment: This work was supported by DFG (FR 1348/31-1), TAB (PlasmidChip PbR 2018 VF 0015), BMBF (NanoWater 02WIL1521, PlasmonBioSense 01DR20010A)
Nanoarchitectures for the monitoring of single molecules and plasmon induced chemical reactions by surface-enhanced Raman scattering (SERS)
Ilko Bald, Kosti Tapio, Sergio Kogikoski Jr., Amr Mostafa, Yuya Kanehira
Institute of Chemistry, University of Potsdam, Potsdam, Germany
DNA origami technology allows for the precise nanoscale assembly of chemical entities that give rise to sophisticated functional materials. We have created a versatile DNA origami nanofork antenna (DONA) by assembling Au or Ag nanoparticle dimers with different gap sizes down to 1.17 nm, enabling signal enhancements in surface-enhanced Raman scattering (SERS) of up to 1011.[1] This allows for single-molecule SERS measurements, which can even be performed with larger gap sizes to accommodate differently sized molecules, at various excitation wavelengths. A general scheme allows to place single analyte molecules into the SERS hot spots using the DNA origami structure exploiting covalent and noncovalent coupling schemes. By using Au and Ag dimers, single-molecule SERS measurements of three dyes and cytochrome c and horseradish peroxidase proteins are demonstrated even under nonresonant excitation conditions, thus providing long photostability during time-series measurement and enabling optical monitoring of single molecules.
Additionally, we show that DNA is able to transfer hot electrons generated by a silver nanoparticle over several nanometers to drive a chemical reaction in a molecule non-adsorbed on the surface.[2] For this we use 8-bromoadenosine introduced in different positions within a double stranded DNA oligonucleotide. The DNA is also used to assemble the nanoparticles into superlattices enabling the use of SERS to track the decomposition reaction. To prove the DNA mediated transfer, the probe molecule was insulated from the charge carriers source, which hindered the reaction. The results indicate that DNA can provide an attractive platform to study the transfer of hot electrons, leading to the future development of more advanced plasmonic catalysts.
1 – Scheme of a DNA origami based nanoantenna (DONA) for single-molecule SERS, which has been used for single protein detection. 2 – Scheme of plasmon induced hot electron transfer along a DNA wire, where a reaction is triggered several nanometers away from the nanoparticle surface.
References:
[1] K. Tapio et al., ACS Nano, (2021) DOI: 10.1021/acsnano.1c00188.
[2] S. Kogikoski Jr. et al., Preprint (2021) DOI: 10.26434/chemrxiv.14114306.v2.
Acknowledgment: This work was supported by the European Research Council (ERC; consolidator grant no. 772752).
One- and two-photon excited SERS for applications in bioanalysis and catalysis: An update
Janina Kneipp, Fani Madzharova, Cecilia Spedalieri, Vesna Živanović, Gergo Peter Szekeres, Daniela Drescher, Zsuzsanna Heiner, Zhiyang Zhang
Department of Chemistry, Humboldt-Universität zu Berlin, Berlin, Germany
We are interested in the new vibrational spectroscopic perspective on biological and chemical processes that we can get from surface-enhanced Raman scattering (SERS), as well as from its two-photon excited analogue, surface-enhanced hyper Raman scattering (SEHRS). We do not consider SERS nor SEHRS a ‘detection tool’, but we use the spectra to better understand the interaction of biological and organic molecules with metal nanostructures, which is the key to successful application of such materials in biotechnology as well as in plasmonic catalysis.
Regarding recent achievements in the former field, we have been able to apply SERS to study biochemical processes in the endolysosomal system of living cells, particularly the influence of specific drugs on enzyme catalysis, [1] and the fragmentation of proteins in vivo. [2] Furthermore, we obtained details on the composition of gold nanostars when they are introduced into living cells, [3] and we provided proof-of-principle that SEHRS in living cells can be obtained under biocompatible excitation conditions in the short-wave infrared range. [4]
SEHRS spectra may also provide useful information when applied for monitoring catalytic reactions, as has been indicated by observation of oxidation products in typical model reactions that are used to study plasmonic catalysis. [5] Here, it may prove helpful to provide a better understanding of the role of plasmonic excitations, e.g., through the generation of hot electrons. Recently, using SERS, we found that the generation of hot carriers can be significantly improved when the right ligands are present at the surface of a metal nanostructure. [6] This has implications for the interpretation of SERS spectra themselves, [7] and for their use in sensing and imaging.
References:
[1] V. Živanović et al., ACS Nano, 13, 9363 (2019)
[2] G.P. Szekeres et al., Analytical Chemistry, 92, 8553 (2020)
[3] C. Spedalieri, Nanoscale, 13, 968 (2021)
[4] Z. Heiner et al., Journal of Raman Spectroscopy, 52, 394 (2020)
[5] F. Madzharova et al., J Phys Chem C., 124 6233 (2020)
[6] Z. Zhang et al., Journal of Catalysis, 383, 153 (2020)
[7] Z. Zhang and J. Kneipp, J. Phys. Chem. Lett. 12, 1542 (2021)
Acknowledgment: This work was supported by ERC Grant 259432, DFG GSC 1013 SALSA, DFG FOR 2177, and Fonds der Chemischen Industrie.
The dynamics of DNA origami filaments growth from an asymmetric monomer
Lena Winat and Barbara Saccà
Center of Medical Biotechnology, University Duisburg-Essen, Essen, Germany
Structural and dynamic features of natural filaments are encoded in the monomeric building unit that is identically repeated along the entire polymer length, suggesting general construction principles for the realization of synthetic biomimetic materials, with stimuli-responsive and self-healing properties.[1-3] A convenient approach to this endeavor relies on the use of DNA structures as the building components of tubules, ribbons or bundles, here collectively referred to as filaments.[4] In this work, we investigate the dynamics of formation of linear polymers made of DNA origami units. Our study builds on a recently reported hierarchical assembly strategy for the generation of different DNA origami filaments from a single asymmetric monomer.[5] Applying base hybridization and base stacking interactions, together with step-wise isolation of assembly intermediates, monomers can be programmed to associate into filaments with different ultrastructures and persistence lengths. Here, we show the impact of such a building principle on the dynamics of filaments growth. We developed a FRET-based assay to monitor the kinetics of tips association and the thermal stability of the resulting filaments, in various design and experimental settings (Fig. 1). Mathematical treatment of the kinetic data according to known polymerization growth models,[6] together with direct observation of polymer formation by atomic force microscopy and dynamic light scattering measurements, show that the rate of filament’s propagation as well as its elongation mechanism are strongly affected by the nature and strength of the interactions occurring at the exposed tips, offering an additional level of control in the programmability of hierarchical DNA origami structures.
Figure 1. Dynamics of DNA origami filaments growth. (a) The kinetics of monomer self-association has been monitored by FRET for different initial monomer concentrations (from ca. 5 nM to 35 nM) and various geometric arrangements (here only the head-to-tail arrangement is represented as an example). (b) The time at which half of the reaction is terminated as a function of the initial monomer concentration evidences the typical nucleation-and-growth signature, whose mechanism is schematically represented in (c).
References:
[1] T. Aida et al. Science, 335, 813-817 (2012).
[2] T. F. de Greef et al. Nature, 453, 171-173 (2008).
[3] L. Brunsveld et al. Chem Rev 101, 4071-4098 (2001).
[4] W. Pfeifer et al. Biol Chem 399, 773-785 (2018).
[5] W. Pfeifer et al. ACS Nano, 12, 44-55 (2018).
[6] A. M. Morris et al. Biochim Biophys Acta, 1794, 375-397 (2009).
Acknowledgment: This work has been realized with financial support from the DFG (CRC 1093, project A6 to B.S. and large instrumentation program INST 20876/336-1 FUGG to B.S.).
Complex metal nanostructures with programmable shapes from simple DNA building blocks
Jingjing Ye (1,2), Olha Aftenieva (3), Türkan Bayrak (2,4), Archa Jain (4), Tobias A. F. König (2,3), Artur Erbe (2,4), and Ralf Seidel (1,2)
(1) Peter Debye Institute for Soft Matter Physics, Universität Leipzig, Linnéstr. 5, 04103 Leipzig, Germany
(2) Center for Advancing Electronics Dresden (cfaed), 01069 Dresden, Germany
(3) Leibniz-Institut für Polymerforschung Dresden e. V., Hohe Straße 6, 01069 Dresden, Germany
(4) Institute of Ion Beam Physics and Materials Research, Helmholtz-Zentrum Dresden-Rossendorf, 01328 Dresden, Germany
Advances in DNA nanotechnology allow to design and fabricate highly complex DNA structures, which uses specific programmable interactions between smaller nucleic acid building blocks. To convey this concept to the fabrication of metallic nanoparticles, an assembly platform was developed based on a few basic DNA structures that can serve as molds. [1,2,3] Programming specific interactions between these elements allowed the assembly of mold superstructures with a range of different geometries(see Figure 1, left). Subsequent seeded growth of gold within the mold cavities enabled the synthesis of complex metal structures including tightly DNA-caged particles, rolling pin- and dumbbellshaped particles as well as T-shaped and loop particles with high continuity. [4] The method further supports the formation of higher-order assemblies of the obtained metal geometries. Based on electrical and optical characterizations (see Figure 1, right), we expect that the developed platform is a valuable tool for a self-assembly-based fabrication of nanoelectronic and nanooptic devices.
Figure 1. Left: Mold elements of the assembly platform. Right: Current-voltage characteristics of the DNA nanolayer (10 nm) measured at different temperatures (4, 100, 200 and 297 K). Inset: SEM image of the contacted DNA nanolayer-nanoelectrode assembly. Arrow mark the DNA nanolayer (red), the protective HSQ resist layer (magenta) and the gold nanoelectrodes (orange). The scale bar is 50 nm.
References:
[1] S. Helmi et al., Nano Letters,14 (11), 6693- 6698 (2014).
[2] T. Bayrak et al., Nano Letters, 18 (3), 2116-2123 (2018).
[3] J. Ye et al., Nano Letters, 19 (4), 2707-2714 (2019).
[4] J. Ye et al., Advanced Materials, 202100381R1 (2021).
Hybridization Chain Reaction based strategies for miRNA detection
Andrea Miti (1), Sophie Thamm (3), Philipp Müller (3), Andrea Csáki (3), Wolfgang Fritzsche (3), Giampaolo Zuccheri (1,2)
(1) Department of Pharmacy and Biotechnology, Alma Mater Studiorum Università di Bologna, Italy.
(2) S3 Center, Institute of Nanoscience of the Italian CNR.
(3) Institute of Photonic Technology, Albert-Einstein-Str. 9, 07745 Jena, Germany
MicroRNAs are attracting great attention for their possible role as biomarkers in diagnosis and follow-up. DNA based biosensors can overcome the common drawbacks faced by the lab-based techniques in miRNA detection. Isothermal amplifications such as Hybridization Chain Reaction (HCR) applied to DNA based sensing, can lead to the improvement of the sensitivity through enhancement of the response [1]. HCR was previously reported also by our group as effective amplification method in the detection of DNA pathogens employing the SPR technology [2]. Target detection and the self-assembly on the surface of metallic nanoparticles can be attained by monitoring the shift in the LSPR peak due to a change in the refractive index at the metal-solution interface [3]. Herein, we report the combination of HCR and a LSPR method based on immobilized gold nanoparticles for the specific detection of microRNAs [4]. We used a specific hairpin-like probe that can efficiently trigger the amplification upon target detection, leading to an improved LOD. The detection method was relatively fast and inexpensive with perspectives in point-of-care applications. We also worked on the implementation of alternative HCR based method that should lead to a higher amplification of the signal. This is based on a DNA triple helix: a “double” HCR can be triggered. Two different recognition elements could be included in the probe on a same single-stranded sequence, allowing a universal assay strategy [5]. We are currently implementing this principle on an electrochemical biosensor.
References:
[1] Dirks, R.M. and N.A. Pierce. PNAS, 2004. 101(43): p. 15275-15278
[2] Spiga, F.M. et al. Biosensors and Bioelectronics54(2014)102–108
[3] a) Jatschka J. et al. Sensing and Bio-Sensing Research. 7 (2016) 62-70. b) Joshi G. K. et al, Nano Lett. 14, (2014), 6955- 6963.
[4] Miti, A. et al. Biosensors and Bioelectronics. 167 (2020) 112465
[5] Zheng, J., et al. Analytical Chemistry, 2014. 86(4): p. 2205-221.
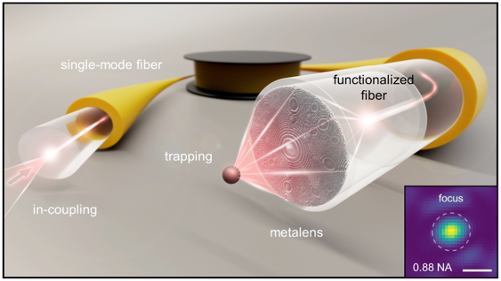
Nanoprinted high-NA meta-lenses on fibers for optical trapping and in-fiber detection of nanoscale species such as SARS-CoV-2
Markus A. Schmidt
Department of Fiber Photonics, Leibniz Institute of Photonic Technologies, Jena, Germany
Hybrid fibers represent a novel class of optical fibers that include sophisticated functionalities on the basis of nano- or microstructures and non-traditional materials. Within this presentation, I will report on our recent results concerning nanostructure-enhanced fibers and single nano-object tracking inside microstructured fibers. The first topic is based on our novel nanostructure implementation approach that is compatible with the fiber geometry (nanoprinting). As applications I will report on optical trapping of micro-beads and bacteria using only one single- mode fiber via the integration of dielectric meta-surfaces, reaching numerical apertures as large as 0.88 [1]. With respect to bio-analysis, I will report on the tracking of single individual nano-objects or of ensembles on nano- objects that diffuse inside various types of microstructured fibers [2, 3]. I will focus on tracking and analysis of ensembles of nano-particles tracking inside hollow-core anti-resonant fibers [4] and will also show our first results on inactivated SARS-CoV-2.
Left: Nanoprinted high-NA meta-lens on modified single mode fiber for optical trapping. Right: Nano-object tracking inside microstructured optical fibers.
The rich world of soft-cavities: strong molecule-cavity coupling and more
Adarsh B Vasista and Bill Barnes
Department of Physics and Astronomy, University of Exeter, United Kingdom
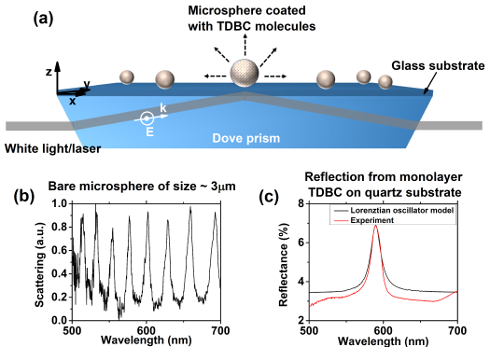
Dielectric microspheres, also called soft cavities, support a rich set of modes depending on the way of excitation, of which Mie scattering resonances and whispering gallery modes (WGMs) are very important. A great advantage of using soft-cavities is that they can be easily trapped and moved in a microfluidic environment. They also show exotic poalrization signatures [1]. Soft-cavities belong to class of open cavities where molecules could be easily adsorbed and desorbed. We are particularly interested in utilizing these modes in coupling molecules in the strong coupling regime.
We study strong-coupling of molecules to the cavity modes (WGMs or Mie modes) of an individual microsphere with J – aggregated cyanine dye molecules at room temperature. We utilize a layer-by-layer (LBL) deposition technique to deposit layers of molecular J – aggregates on the microsphere surface. The coated microspheres were probed using evanescent excitation based dark-field microscopy(see figure 1). The evolution of the Rabi splitting as a function of the number of deposited layers is also studied. The results are analyzed using a coupled oscillator model, and using finite element method (FEM) numerical simulations [2]. Further we have probed molecule-cavity coupling with Mie resonances by exciting the microsphere through far-field excitation.
Figure1: (a) Schematic representing the J-aggregate PDAC/TDBC coated microsphere system. Individual microspheres were probed using evanescent excitation based dark field spectroscopy. (b) Typical dark field scattering spectrum collected from a microsphere of size ∼3 μm placed on glass substrate showing spectrally sharp whispering gallery modes. (c) Experimentally measured reflectance spectrum of a monolayer PDAC/TDBC on quartz substrate in comparison with that calculated using Lorenztian oscillator model.
References:
[1] Vasista, A. B., et al., Advanced Optical Materials, 6(22), 1801025 (2018).
[2] Vasista A.B., et al., Nano Lett. 20 (3) 1766-1773 (2020).
Acknowledgment: This work was supported by ERC through PhotMat project (ERC-2016-AdG-742222 www.photmat.eu).
© 2025 · Biophotonics4Future
Leibniz Institute of Photonic Technology
Albert-Einstein-Str. 9
07745 Jena | Germany
www.leibniz-ipht.de